The application of Next Generation Sequencing in Cancer Diagnosis and Treatment
ONE BY ONE TESTING APPROACH vs ALL AT ONCE
INTRODUCTION
Next Generation Sequencing and Cancer
In the current timeline, a series of high-throughput molecular tests, such as Fragment analysis, Sanger sequencing, Real time PCR, Real time multiplexed PCR are well known for mutation detection.
However, there is a critical need for incorporating Next generation sequencing (NGS) into routine clinical practice. In this newsletter, we aim to review the advantages of NGS for the detection of mutation and personalized medicine along with its limitations.
Cancer prevalence in India
The current Indian population is 1,270,272,105 (1.27 billion). The incidence of cancer in India is 70-90 per 100,000 population and cancer prevalence is established to be around 2,500,000 (2.5 million) with over 800,000 new cases and 5, 50,000 deaths occurring each year. More than 70% of the cases present in advanced stage account for poor survival and high mortality. About 6% of all deaths in India are due to cancers which contribute to 8% of global cancer mortality.
According to Indian Council of Medical Research (ICMR) data on site specific cancer burden, in males, the most common are cancers of mouth/pharynx, esophagus, stomach, lung/bronchi while as in females, the common cancers are cervix, breast, mouth/oropharynx and esophagus
(Courtesy: http://www.dailyexcelsior.com/cancer-scenario-india/).
Based on GLOBOCAN estimates, about 14.1 million new cancer cases and 8.2 million deaths occurred in 2012 worldwide. Over the years, the burden has shifted to less developed countries, which currently account for about 57% of cases and 65% of cancer deaths worldwide. Lung cancer is the leading cause of cancer death among males in both more and less developed countries and has surpassed breast cancer as the leading cause of cancer death among females in more developed countries. Breast cancer remains the leading cause of cancer death among females in less developed countries (Lindsey A. Torre et al. 2015).
What causes Cancer?
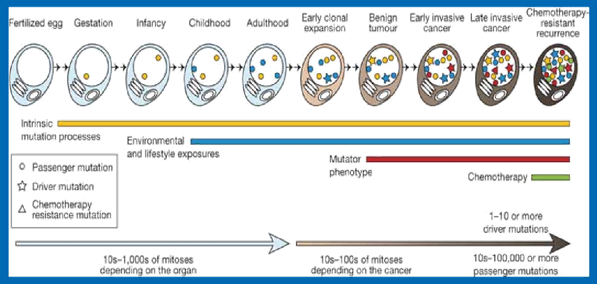
Cancers are a large family of diseases that involve abnormal cell growth with the potential to invade to other parts of the body. They form a subset of Neoplasms. A neoplasm or tumor is a group of cells that have undergone unregulated growth and will often form a mass or lump, but may be distributed diffusely. Cancer is a class of diseases characterized by out-of-control cell growth. As the cells divide during the lifetime of an individual, certain changes or mutations occur in the DNA. These changes or mutations keep on accumulating in the cell and may cross the threshold limit. Once this threshold limit is crossed, the cell division may go haywire and result in cancer test.
There are over 100 different types of cancer and each is classified by the type of cell that is initially affected. More dangerous or malignant tumors form when two things occur:
- A cancerous cell manages to move throughout the body using the blood or lymphatic systems, destroying a healthy tissue in a process called Invasion.
- That cell manages to divide and grow, making new blood vessels to feed itself in a process called Angiogenesis.
What is Mutation?
A gene mutation is a permanent alteration in the DNA sequence that makes up a gene, such that the sequence differs from what is found in most people. Mutations range in size, they can affect anywhere from a single DNA building block (base pair) to a large segment of a Chromosome that includes multiple genes.
Gene mutations can be classified in two major ways:
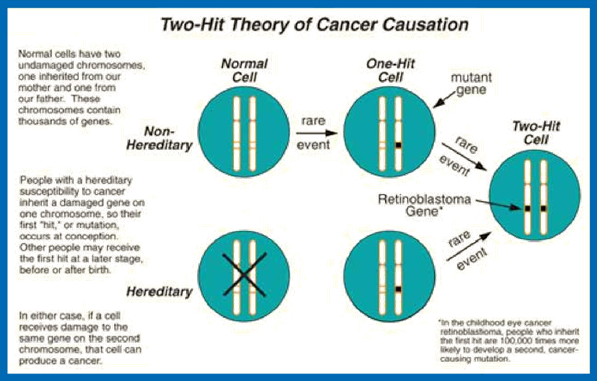
- Hereditary mutations are inherited from a parent and are present throughout a person’s life in virtually every cell in the body. These mutations are also called Germline mutations because they are present in the parent’s egg or sperm cells, which are also called germ cells.
- Acquired (or Somatic) mutations occur at some time during a person’s life and are present only in certain cells, not in every cell in the body. These changes can be caused by environmental factors such as ultraviolet radiation from the sun or can occur if a mistake is made as DNA copies itself during cell division. Acquired mutations in somatic cells (cells other than sperm and egg cells) cannot be passed on to the next generation.
Knudson’s “Two-Hit” Theory of cancer Causation
(Courtesy: http://www.fccc.edu/research/areas/advisors/knudson/twoHit.html)
Example: Single Nucleotide Polymorphism (SNP)
Single Nucleotide Polymorphism or SNP is a type of genetic variation, where a single letter difference occurs in the DNA sequence of an individual, when compared to others.
Example: Sequence 1: —-AGCCTAATGGGC—-
Sequence 2: —-AGCCTAAGGGGC—-
Here, in the given example, the first sequence differs from the second sequence only by a single letter (nucleotideT/G). This single letter variation affects many phenotypic traits, disease susceptibility/ resistance, response to drugs, chemicals, radiation etc.
Types of cancer and known affected genes
| Type of cancer | Mutated Genes responsible for cancer | |
| Lung cancer | AKT, ALK,BRAF, EGFR, ERBB2, FGFR, KRAS, NRAS, PIK3CA, PTEN, ROS1, RET | |
| Colorectal cancer | AKT, BRAF, KRAS, NRAS, PIK3CA, PTEN, SMAD4 | |
| Melanoma | BRAF, CTNNB1, KIT, MAP2K, NRAS, GNA11,GNAQ | |
| Ovarian cancer | BRAF, KRAS, PI3CA, PTEN | |
Options for Cancer Diagnosis-NGS
- Genome-wide association studies and cancer genome sequencing have identified ~ 400 genes which can go kaput in cancer (100 Oncogenes, 30 Tumor-Specific Genes, Several genome repair genes, Chromatin remodelling genes).
- List out driver mutations in a tumor type from an organ. Check for each molecular alteration one by one or the currently druggable ones simultaneously but in a separate experiment for each
Single gene assays.
- Check out for all drivers in an organ specific cancer simultaneously by Multiplexed PCR followed by Sanger’s sequencing or multiplexed RT- PCR or Snapshot assays.
- Significantly Mutated Genes (SMG) = 50 such genes harbour most cancer gene alterations. High Confidence Drivers (HCD). Likely that one or more of these is altered in any Cancer type. Look for all the SMG (HCD) together using NGS.
- Knowing what mutation or alteration caused the cancer can help in targeted therapy, thus reducing overall costs and improving the outcome of the therapy.
Why Next Gen Sequencing?
Next Generation sequencing (NGS) is one of the most significant technological advances in the biological sciences of the last 30 years. Targeted DNA enrichment methods allow even higher genome throughout at a reduced cost per sample. NGS is now maturing to the point where it is being considered by many laboratories for routine diagnostic use. The sensitivity, speed and reduced cost per sample make it a highly attractive platform compared to other sequencing modalities. Moreover, as we identify more genetic determinants of cancer, there is a greater need to adopt multi-gene assays that can quickly and reliably sequence complete genes from individual patient samples.
There are various methods to know these driver mutations – Sanger sequencing, Real time PCR, Next Gen Sequencing etc.
Comparative view of Sanger sequencing, Real time sequencing and Next Generation Sequencing
| Sanger Sequencing | Real time PCR | Next gen sequencing | ||
| • This requires at least | • The sensitivity is better | • | Highly sensitive assay | |
| 10-20% tumor cells | than Sanger sequencing. | can detect mutations in | ||
| to be present, otherwise | • Each assay is highly | samples having 5% | ||
| the mutation is lost in | targeted and can detect | tumor cells. | ||
| the background DNA | only one mutation in | • | Can be multiplexed. | |
| which comes from | most cases. | • Many genes can be | ||
| normal tissue. | • Does not detect | targeted at the same | ||
| • This method can detect | neighboring mutations. | time, thus detecting | ||
| neighbouring mutations. | many mutations in the | |||
| • Each mutation site is | same assay. | |||
| • Each DNA nucleotide is | ||||
| analyzed only once. | ||||
| read many times | ||||
| (100-1000 times) which | ||||
| adds to the specificity of | ||||
| the assay. | ||||
Genes covered in 50 Gene Cancer panel
| Next Generation | Sample preparation workflow | Sequencing | ||||||
Principle of NGS technique
| Chip-based semi-conductor sequencing | Next Generation Sequencing | |
| data analysis workflow | ||
Advantages of NGS practice in SMG than normal sequencing diagnosis
The tumor tissue which is extracted out, needs to undergo multiple diagnostic assays – Histopathology, IHC, Molecular and Cytogenetic tests. Thus, the sample is a very precious sample. If subjected to multiple tests one by one, the tissue becomes a limiting factor. It is thus advisable to go for all the molecular tests in one go rather than sequentially.
- NGS is Helpful for biopsy analysis: From a single biopsy, ≤50 genes are impossible to analyze using Single gene assay.
- Min of 10ng of DNA is required for a single NGS reaction (Samuel A. Yousem et al.,2013): ≥
10ng of DNA is required for a single sequencing reaction using Sanger’s method.
- One time test can screen 50 genes (SMG): Sanger’s sequencing needs lot of time and DNA to complete 50 genes for cancer diagnosis.
- Test can be performed in Formalin fixed tissues: Advanced design of Primer sequence allows to cover the targeted part of the gene using overlapping technology. In this technique, short fragmented DNA can be amplified and overlapped to find the whole gene sequence. However, using Sanger’s sequencing, a short stretch of DNA can be analyzed at a time.
- Quantitative measurement of mutation: Quantitative measurement of mutation can be declared accurately. In the Sanger’s method, accurate quantitative detection of the mutation is not possible.
- Cancer needs very accurate and immediate diagnosis: In case of routine sequencing, time is the major fact which is prolonged. This can leads to early tumor to late stage cancer diagnosis.
The answer – Why go sequentially ( For example, if lung cancer status
of EGFR/KRAS/ NRAS/ALK/PTEN etc. needs to be known) when you can have the answer in less time and approximately the same cost, if you do a 50
Gene Cancer panel.
Types of sample used
- Biopsy
- Formalin fixed tissue or paraffin embedded tissue block
Limitations
- Cannot identify large insertions/ deletions
- CNV – not always
- Cannot identify translocations
- Identify mutations – no data about sensitivity to a drug (clinical utility not fully established)
- Requires complex bioinformatics
Case Study: Unusual EGFR mutation and
its effect in drug response-Salivary gland carcinomas
Activating mutations within the Epidermal Growth Factor Receptor (EGFR) tyrosine kinase domain identify non-small cell lung cancer patients with improved clinical response to tyrosine kinase inhibitor therapy.
EGFR mutations in the tyrosine kinase domain is a rare event in Salivary Gland Carcinomas (SGC). However, R Dashe and his group identified that EGFR mutation screening was helpful for SGC.
In his study, he had screened 25 SGC samples for the EGFR mutation. Out of 25 samples, surprisingly two samples were found to have Exon 19 deletion. Exon 19 deletion detection increases the chance of positive response to Gefitinib therapy. (Zhang et al., 2014; Jackman et al., 2006)
Cancer genomes are riddled with mutations. The challenge is of efficiently picking out the guilty drivers in the huge identification parade.
The impact of Cancer in India is far greater than mere number of cancer cases. Its diagnosis causes immense emotional trauma and its treatment has a major economic burden. The initial diagnosis of cancer is perceived by many patients as a grave event with more than one-third of them suffering from anxiety and depression.
Treatment facilities are also mostly limited to urban areas of the country. There is no uniform protocol for management and the availability and affordability of cancer treatment shows wide disparities. The majority of patients with cancer present to a cancer treatment centre in late stages of the disease (80% are advanced) and this adds to the already high morbidity, mortality and expenditure.
Treatment results are about 20% less than what is observed for similar conditions in more developed countries, mostly due to late diagnosis and inappropriate treatment. Pediatric cancers are highly curable but this has not been achieved in India due to lack of access to quality care and lack of support systems.
Cancer care needs palliative care, finances and coordination in the accurate timing.
Test range available
Test code : N097
Test Name : CANCER 50 GENE PANEL
Method: NEXT GENERATION SEQUENCING
Specimen: FORMALIN FIXED PARAFFIN EMBEDDED TISSUE BLOCK FROM TUMOUR SITE *Report: SAMPLE BY 1ST / 15TH OF EVERY MONTH REPORT 10 WORKING DAYS
* In conjunction with Rajiv Gandhi Cancer Hospital & Research centre -Rohini
References
- Rothberg et al., Nature. 2011 Jul 20;475(7356):348-52. doi: 10.1038/nature10242.An integrated semiconductor device enabling non-optical genome sequencing.
- Lindsey A. Torre et al., CA CANCER J CLIN 2015;65:87–108.doi: 10.3322/caac.21262. Global cancer statistics, 2012.
- Samuel A. Yousem et al., January 2013, doi:10.1378/chest.12-1917. Pulmonary Langerhans Cell Histiocytosis:Profiling of Multifocal Tumors Using Next-Generation Sequencing Identifies Concordant Occurrence of BRAF V600E Mutations.
- Michael M. H. Yang et al., May 2012, doi:10.3171/2012.1.PEDS11326. Clinical Validation of KRAS, BRAF, and EGFR Mutation Detection Using Next-Generation Sequencing.
- http://www.dailyexcelsior.com/cancer-scenario-india/)
- http://www.fccc.edu/research/areas/advisors/knudson/twoHit.html
- R Dahse et al., Br J Cancer. 2009 Feb 24; 100(4): 623–625. doi: 10.1038/sj.bjc.6604875.Epidermal growth factor receptor kinase domain mutations are rare in salivary gland carcinomas.
- David M. Jackman et al., Clin Cancer Res July 1, 2006 12;3908. doi: 10.1158/1078-0432.Exon 19 Deletion Mutations of Epidermal Growth Factor Receptor Are Associated with Prolonged Survival in Non–Small Cell Lung Cancer Patients Treated with Gefitinib or Erlotinib.
- Yaxiong Zhang et al., PLoS One. 2014; 9(9): e107161.doi: 10.1371/journal.pone.0107161Patients with Exon 19 Deletion Were Associated with Longer Progression-Free Survival Compared to Those with L858R Mutation after First-Line EGFR-TKIs for Advanced Non-Small Cell Lung Cancer: A Meta-Analysis