The application of Next Generation Sequencing(NGS) in Cancer Diagnostics and Treatment
ONE BY ONE TESTING APPROACH VS ALL AT ONCE
What Causes Cancer? – A genomic perspective
- As the cells divide during the lifetime of an individual, certain changes or mutations occur in the DNA
- These changes or mutations keep on accumulating in the cell and may cross the threshold limit.
- Once this threshold limit is crossed, the cell division may go haywire and result in cancer
How to treat the cancer?
- One of the approach to treat cancer is to identify the change or the mutation in the DNA which caused the cancer and then to provide an antidote for it.
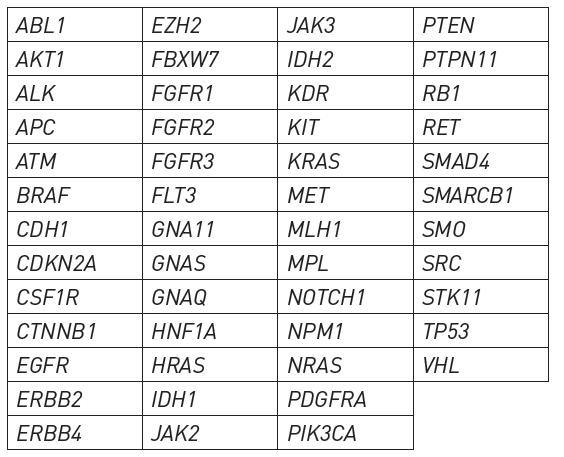
- By comparing the healthy individuals with the patients, scientists have identified ~ 400 genes which go kaput in cancer. (100 oncogenes, 30 Tumour Specific Genes, several genome repair genes, chromatin remodelling genes).
- Out these 400, Significantly Mutated Genes (SMG) are 50 such genes which harbour most cancer gene alterations. These are High Confidence Drivers (HCD). It is Likely that one or more of these is altered in any cancer type
- Knowing what mutation or alteration caused the cancer can help in targeted therapy, thus reducing overall costs and improving the outcome of the therapy.
Options for Diagnosis
- List out driver mutations in a tumor type from an organ. Check for each molecular alterations one by one or the currently drug targeted ones simultaneously but in a separate experiment for each- Single gene assays.
- Check out for all drivers in an organ specific cancer simultaneously by Multiplexed PCR followed by Sanger’s sequencing or multiplexed RT- PCR or Snapshot assays.
- Look for all the SMG (HCD) together using NGS.
Why Next Gen Sequencing
There are various methods to “know” these driver mutations – Sanger sequencing, Real Time PCR, Next Generation Sequencing etc.
Technique
- Sanger Sequencing – requires at least 10-20% tumor cells to be present, otherwise the mutation is “lost” in the background DNA which comes from normal tissue. Can detect neighboring mutations. Each mutation site is analyzed only once
- Real time PCR – the sensitivity is better than Sanger sequencing, but each assay is highly targeted and can detect only one mutation in most cases. Does not detect neighboring mutations
- Next gen sequencing – Highly sensitive assay, can detect mutations in samples having 5% tumor cells. Can be multiplexed, so many genes can be targeted at the same time, thus detecting many mutations in the same assay. Each DNA nucleotide is read many times (100-1000 times) which adds to the specificity of the assay
Sample
The tumor tissue which is extracted out needs to undergo multiple diagnostic assays – histopathology, IHC, molecular and cytogenetic tests. Thus the sample is a very precious sample. If subjected to multiple tests one by one the tissue becomes a limiting factor. It is thus advisable to go for all the molecular tests in one go rather than sequentially.
The percentage of tumor cells is often low. Thus a sensitive technique is required to detect these mutations in a background of wild type cellular DNA. NGS uses massively parallel sequencing wherein high degree of accuracy is achieved by sequencing the same base again and again.
The answer – why go sequentially ( for e.g. for lung cancer status of EGFR/KRAS/NRAS/ALK/PTEN etc need to be known) when you can have the answer in less time and approx. the same cost if you do a 50 Gene Cancer Panel.
Genes covered